
| Name: AGTPBP1 | Sequence: fasta or formatted (1186aa) | NCBI GI: 170763513 | |
|
Description: ATP/GTP binding protein 1
|
Referenced in:
| ||
|
Composition:
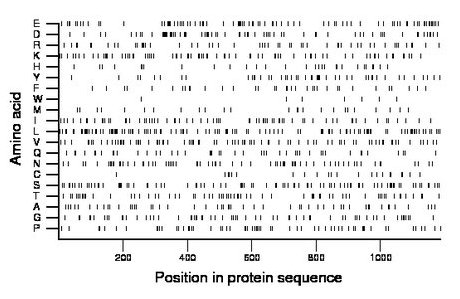
Amino acid Percentage Count Longest homopolymer A alanine 4.5 53 2 C cysteine 1.9 23 2 D aspartate 6.2 73 2 E glutamate 7.2 85 2 F phenylalanine 3.0 35 1 G glycine 5.4 64 3 H histidine 2.5 30 1 I isoleucine 5.8 69 2 K lysine 6.5 77 2 L leucine 9.9 117 2 M methionine 2.2 26 1 N asparagine 6.0 71 2 P proline 5.1 61 3 Q glutamine 4.0 47 3 R arginine 4.5 53 2 S serine 8.0 95 2 T threonine 5.6 67 2 V valine 7.0 83 3 W tryptophan 0.7 8 2 Y tyrosine 4.1 49 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 ATP/GTP binding protein 1 AGBL1 0.289 ATP/GTP binding protein-like 1 AGBL3 0.139 carboxypeptidase 3, cytosolic AGBL2 0.130 carboxypeptidase 2, cytosolic AGBL4 0.077 ATP/GTP binding protein-like 4 AGBL5 0.052 ATP/GTP binding protein-like 5 isoform 1 AGBL5 0.052 ATP/GTP binding protein-like 5 isoform 3 ZBTB7C 0.005 zinc finger and BTB domain containing 7C BAZ2B 0.005 bromodomain adjacent to zinc finger domain, 2B [Homo... UPF2 0.004 UPF2 regulator of nonsense transcripts homolog [Homo... UPF2 0.004 UPF2 regulator of nonsense transcripts homolog [Homo... UBLCP1 0.004 ubiquitin-like domain containing CTD phosphatase 1 ... NRD1 0.004 nardilysin isoform b NRD1 0.004 nardilysin isoform a CEBPZ 0.003 CCAAT/enhancer binding protein zeta ZNF638 0.003 zinc finger protein 638 ZNF638 0.003 zinc finger protein 638 KIF21A 0.003 kinesin family member 21A ARID4B 0.003 AT rich interactive domain 4B isoform 1 LOC100287665 0.003 PREDICTED: hypothetical protein XP_002342862 COL12A1 0.003 collagen, type XII, alpha 1 long isoform precursor [... C12orf70 0.003 hypothetical protein LOC341346 LOC646817 0.003 PREDICTED: similar to SET translocation LOC646817 0.003 PREDICTED: similar to SET translocation LOC646817 0.003 PREDICTED: similar to SET translocation CPA4 0.003 carboxypeptidase A4 preproprotein C4orf8 0.003 hypothetical protein LOC8603 HAP1 0.003 huntingtin-associated protein 1 isoform 4 HAP1 0.003 huntingtin-associated protein 1 isoform 3 HAP1 0.003 huntingtin-associated protein 1 isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
