
| Name: AGBL2 | Sequence: fasta or formatted (902aa) | NCBI GI: 31657119 | |
|
Description: carboxypeptidase 2, cytosolic
|
Referenced in:
| ||
|
Composition:
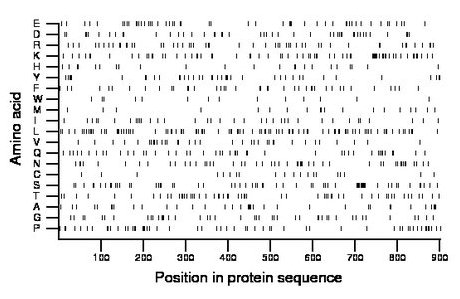
Amino acid Percentage Count Longest homopolymer A alanine 4.1 37 3 C cysteine 2.2 20 1 D aspartate 5.0 45 2 E glutamate 5.5 50 2 F phenylalanine 4.7 42 2 G glycine 5.0 45 2 H histidine 2.7 24 2 I isoleucine 4.0 36 1 K lysine 7.8 70 6 L leucine 10.6 96 2 M methionine 2.2 20 1 N asparagine 5.8 52 4 P proline 6.3 57 4 Q glutamine 5.1 46 2 R arginine 5.7 51 2 S serine 7.3 66 2 T threonine 6.1 55 1 V valine 4.1 37 2 W tryptophan 1.6 14 1 Y tyrosine 4.3 39 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 carboxypeptidase 2, cytosolic AGBL3 0.367 carboxypeptidase 3, cytosolic AGTPBP1 0.166 ATP/GTP binding protein 1 AGBL1 0.157 ATP/GTP binding protein-like 1 AGBL4 0.113 ATP/GTP binding protein-like 4 AGBL5 0.061 ATP/GTP binding protein-like 5 isoform 1 AGBL5 0.061 ATP/GTP binding protein-like 5 isoform 3 ZNF573 0.011 zinc finger protein 573 BRCA2 0.009 breast cancer 2, early onset CIR1 0.008 CBF1 interacting corepressor CEP350 0.007 centrosome-associated protein 350 LOC100133698 0.006 PREDICTED: hypothetical protein LOC100292671 0.006 PREDICTED: similar to CYorf16 protein LOC100290384 0.006 PREDICTED: hypothetical protein XP_002348288 LOC100288465 0.006 PREDICTED: hypothetical protein XP_002343910 C14orf102 0.006 hypothetical protein LOC55051 isoform 1 ESF1 0.006 ABT1-associated protein CEP290 0.005 centrosomal protein 290kDa PCF11 0.005 pre-mRNA cleavage complex II protein Pcf11 PRG4 0.005 proteoglycan 4 isoform A TAF1D 0.005 TATA box binding protein (TBP)-associated factor, RN... LOC100294364 0.005 PREDICTED: hypothetical protein PPIG 0.005 peptidylprolyl isomerase G RSBN1L 0.005 round spermatid basic protein 1-like PASD1 0.005 PAS domain containing 1 ZNF654 0.005 zinc finger protein 654 MOG 0.005 myelin oligodendrocyte glycoprotein isoform beta4 pr... MOG 0.005 myelin oligodendrocyte glycoprotein isoform alpha4 p... BCLAF1 0.005 BCL2-associated transcription factor 1 isoform 3 [H... BCLAF1 0.005 BCL2-associated transcription factor 1 isoform 1 [Hom...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
