Polymorphism
The human genome has considerable polymorphism when different individuals or the alleles of an individual are compared. Some of this variation results in obviously deleterious mutations, whereas other differences may lack any clear phenotype. Unusual levels of polymorphism are often indicators of the selective pressures acting on the genome.Although most of the polymorphism described in the Guide is related to coding regions of genes, there are many well-known examples that affect gene regulation and splicing. One of the best described is related to persistence of lactase (LCT) expression in adults associated with milk tolerance. Many mutations affecting splicing are known for the globin genes. Several classes of polymorphism that directly or indirectly affect coding sequences are presented in this section.
Coding sequence variation
The HLA genes are notable for the large number of alleles found in the population. The receptors on NK cells (see Additional Immunoglobulin-related Receptors) that interact with HLA proteins also display considerable polymorphism.The ABO locus, controlling the main blood groups (see Hematopoiesis and Erythrocytes), is a case not only of frequent polymorphism with a common null allele but of an enzyme where the natural variants provide extensive functional information.
Several common disease loci are associated with resistance to infection. Among these are G6PD (glucose-6-phosphate dehydrogenase; also isoform), CFTR (cystic fibrosis conductance regulator), and HBB (hemoglobin β). The alterations in iron storage seen with mutations in HFE (hemochromatosis; also many isoforms) might impact bacterial infections. Mutant alleles of MEFV (pyrin) might represent selection for enhanced response to certain pathogens. Also of note is the resistance to HIV associated with mutation at CCR5 (also alt mRNA).
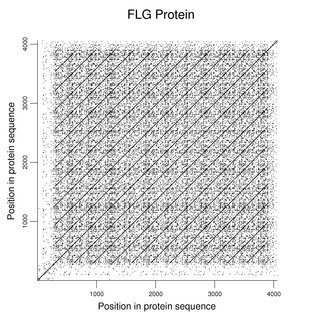
Mutations of filaggrin (FLG) affect the granule layer of the skin and are common in certain populations. As shown in the following dot plot (word size 3), most of this relatively large protein consists of a large number of repeating regions that yield mature filaggrin after proteolytic cleavage. The reference sequence has 11 repeats and some variation in repeat number is known. Because the individual repeats have function, mutations are often frameshifts or produce nonsense codons.
Polymorphism in the N-acetyltransferases has been studied intensively because of the role of the enzymes in the metabolism of xenobiotics. A number of the cytochrome P450 enzymes have been studied for similar reasons.
Variation in gene number
Duplication and divergence is a major mechanism for the evolution of new functions. Examples of loci with common variation in gene number includes amylase (see Hexokinases and Initial Sugar Metabolism) and complement protein 4 (see Complement). Note the variation in the number of copies of CCL3L1 (see Chemokines and Their Receptors), a ligand for CCR5.Variation in the number of DAZ genes on the Y chromosome (see Testes and Sperm) has been investigated because of the association of variations at this locus with fertility. Variation in gene content is also seen for the receptors on NK cells mentioned above.
Pathway analysis
In a number of pathways presented in the text, human mutations have been observed at most or all steps, facilitating greater understanding of the roles of individual genes. Examples include glycolysis, the pentose pathway, glycogen metabolism, the urea cycle, heme synthesis, and the complement pathway. Mutations affecting pigmentation are described in the section on skin.Human mutations are known in many proteins involved in the biogenesis of lysosome-related organelles. The text describes how mouse mutants supplement the information derived from known human variation.
Unstable loci
Proteins with low-complexity regions (see Protein Composition and Structure) have frequent polymorphisms. Notable examples of this type include the polyglutamine tracts in ATXN1 (also alt mRNA) and ATXN2 (see Cerebellum) and polyalanine tracts in members of the FOX family.Extraordinary levels of polymorphism are seen with the LPA gene (see Lipoproteins). In this case, the variation derives from the number of copies of a 114-aa repeating unit that makes up most of the protein. The reference allele has 16 copies of the repeat.
New mutations are frequently seen at the very large locus encoding DMD (dystrophin, with many alt mRNAs and isoforms). Alterations in many of the largest genes in the genome are observed in tumors.
Notes and references
Many references and other information for individual genes can be found in the RefSeq entries linked via the pages for the proteins mentioned in this section. A table of these entries (with the corresponding gene identifiers) and a collection of their sequences also are available.DMD and HFE have many alternate products that can be accessed via the links in this section.
See also the additional reading for this chapter.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.