
| Name: APEH | Sequence: fasta or formatted (732aa) | NCBI GI: 23510451 | |
|
Description: N-acylaminoacyl-peptide hydrolase
|
Referenced in:
| ||
|
Composition:
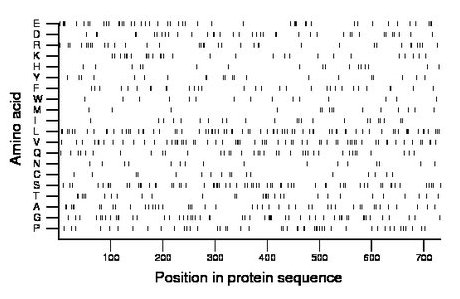
Amino acid Percentage Count Longest homopolymer A alanine 6.1 45 3 C cysteine 2.3 17 1 D aspartate 5.3 39 2 E glutamate 6.4 47 2 F phenylalanine 4.2 31 2 G glycine 7.8 57 2 H histidine 2.5 18 2 I isoleucine 3.1 23 1 K lysine 3.8 28 2 L leucine 10.5 77 3 M methionine 2.2 16 1 N asparagine 2.2 16 1 P proline 6.6 48 3 Q glutamine 5.1 37 1 R arginine 4.8 35 2 S serine 8.7 64 2 T threonine 4.0 29 2 V valine 9.2 67 3 W tryptophan 2.2 16 2 Y tyrosine 3.0 22 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 N-acylaminoacyl-peptide hydrolase DPP9 0.032 dipeptidylpeptidase 9 DPP8 0.021 dipeptidyl peptidase 8 isoform 3 DPP8 0.021 dipeptidyl peptidase 8 isoform 1 FAP 0.019 fibroblast activation protein, alpha subunit DPP6 0.018 dipeptidyl-peptidase 6 isoform 3 DPP6 0.018 dipeptidyl-peptidase 6 isoform 2 DPP6 0.018 dipeptidyl-peptidase 6 isoform 1 DPP4 0.008 dipeptidylpeptidase IV C20orf135 0.007 hypothetical protein LOC140701 DPP10 0.007 dipeptidyl peptidase 10 isoform short DPP10 0.007 dipeptidyl peptidase 10 isoform long PREP 0.007 prolyl endopeptidase CACNA1A 0.006 calcium channel, alpha 1A subunit isoform 4 CES3 0.006 carboxylesterase 3 TIAM2 0.005 T-cell lymphoma invasion and metastasis 2 isoform a ... PGA4 0.005 pepsinogen 4, group I PGA3 0.005 pepsinogen 3, group I PGA5 0.005 pepsinogen 5, group I HERC1 0.005 guanine nucleotide exchange factor p532 LOC100134348 0.005 PREDICTED: similar to TBC1 domain family, member 3 ... LOC100293189 0.005 PREDICTED: hypothetical protein LOC100291266 0.005 PREDICTED: hypothetical protein XP_002346666 LOC100288943 0.005 PREDICTED: hypothetical protein XP_002342523 PDCD4 0.004 programmed cell death 4 isoform 2 PDCD4 0.004 programmed cell death 4 isoform 1 PGK2 0.003 phosphoglycerate kinase 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
