
| Name: LOC100293189 | Sequence: fasta or formatted (140aa) | NCBI GI: 239753870 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
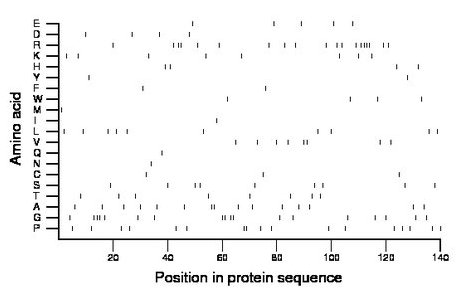
Amino acid Percentage Count Longest homopolymer A alanine 10.7 15 2 C cysteine 2.1 3 1 D aspartate 2.9 4 1 E glutamate 3.6 5 1 F phenylalanine 1.4 2 1 G glycine 12.9 18 3 H histidine 2.9 4 1 I isoleucine 0.7 1 1 K lysine 5.7 8 1 L leucine 7.1 10 1 M methionine 0.7 1 1 N asparagine 0.7 1 1 P proline 12.1 17 2 Q glutamine 0.7 1 1 R arginine 13.6 19 4 S serine 6.4 9 1 T threonine 5.7 8 1 V valine 5.7 8 2 W tryptophan 2.9 4 1 Y tyrosine 1.4 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291266 1.000 PREDICTED: hypothetical protein XP_002346666 LOC100288943 1.000 PREDICTED: hypothetical protein XP_002342523 FHDC1 0.029 FH2 domain containing 1 AHDC1 0.029 AT hook, DNA binding motif, containing 1 APEH 0.025 N-acylaminoacyl-peptide hydrolase RASSF5 0.022 Ras association (RalGDS/AF-6) domain family 5 isofor... RASSF5 0.022 Ras association (RalGDS/AF-6) domain family 5 isofor... LOC729580 0.022 PREDICTED: hypothetical protein LOC729580 0.022 PREDICTED: hypothetical protein LOC729580 0.022 PREDICTED: hypothetical protein TRIO 0.018 triple functional domain (PTPRF interacting) ACIN1 0.018 apoptotic chromatin condensation inducer 1 BZRAP1 0.018 peripheral benzodiazepine receptor-associated prote... BZRAP1 0.018 peripheral benzodiazepine receptor-associated prote... BAHD1 0.018 bromo adjacent homology domain containing 1 MEPCE 0.018 bin3, bicoid-interacting 3 LOC100130128 0.015 PREDICTED: hypothetical protein LOC100130128 0.015 PREDICTED: hypothetical protein LOC100130128 0.015 PREDICTED: hypothetical protein SRRM2 0.015 splicing coactivator subunit SRm300 LOC100293466 0.015 PREDICTED: hypothetical protein LOC100291878 0.015 PREDICTED: similar to RIKEN cDNA C230030N03 LOC100287213 0.015 PREDICTED: hypothetical protein LOC100287213 0.015 PREDICTED: hypothetical protein XP_002343128 OBSL1 0.011 obscurin-like 1 LOC729590 0.011 PREDICTED: hypothetical protein LOC728767 0.011 PREDICTED: hypothetical protein LOC729590 0.011 PREDICTED: hypothetical protein LOC728767 0.011 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
