
| Name: FAM190B | Sequence: fasta or formatted (834aa) | NCBI GI: 80861486 | |
|
Description: granule cell antiserum positive 14
|
Referenced in:
| ||
|
Composition:
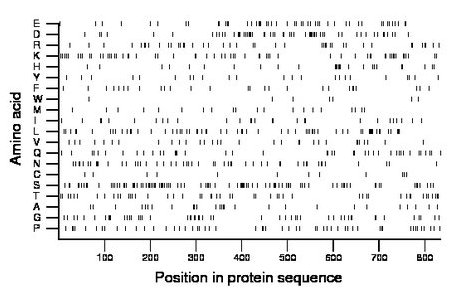
Amino acid Percentage Count Longest homopolymer A alanine 3.2 27 2 C cysteine 1.4 12 1 D aspartate 6.8 57 3 E glutamate 6.4 53 2 F phenylalanine 3.2 27 2 G glycine 6.1 51 2 H histidine 3.2 27 3 I isoleucine 3.4 28 2 K lysine 7.2 60 3 L leucine 7.7 64 2 M methionine 2.5 21 1 N asparagine 6.1 51 2 P proline 6.7 56 2 Q glutamine 5.0 42 2 R arginine 5.0 42 2 S serine 12.6 105 3 T threonine 6.0 50 2 V valine 3.7 31 1 W tryptophan 0.8 7 1 Y tyrosine 2.8 23 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 granule cell antiserum positive 14 FAM190A 0.040 KIAA1680 protein isoform 1 FAM190A 0.033 KIAA1680 protein isoform 2 DSPP 0.014 dentin sialophosphoprotein preproprotein RSF1 0.009 remodeling and spacing factor 1 RPGRIP1 0.008 retinitis pigmentosa GTPase regulator interacting p... SPANXN2 0.008 SPANX-N2 protein ARID1B 0.008 AT rich interactive domain 1B (SWI1-like) isoform 1 ... NUFIP2 0.007 nuclear fragile X mental retardation protein interac... KIAA1430 0.007 hypothetical protein LOC57587 ACRC 0.007 ACRC protein BAZ2B 0.007 bromodomain adjacent to zinc finger domain, 2B [Homo... NAV3 0.007 neuron navigator 3 PALM2 0.006 paralemmin 2 isoform b PALM2 0.006 paralemmin 2 isoform a PALM2-AKAP2 0.006 PALM2-AKAP2 protein isoform 2 PALM2-AKAP2 0.006 PALM2-AKAP2 protein isoform 1 EDEM3 0.006 ER degradation enhancer, mannosidase alpha-like 3 [... TMEM200A 0.006 transmembrane protein 200A CYLC1 0.006 cylicin, basic protein of sperm head cytoskeleton 1... ETAA1 0.006 ETAA16 protein ESF1 0.006 ABT1-associated protein RBM12B 0.005 RNA binding motif protein 12B TTLL7 0.005 tubulin tyrosine ligase-like family, member 7 SSFA2 0.005 sperm specific antigen 2 isoform 1 SSFA2 0.005 sperm specific antigen 2 isoform 2 MACROD2 0.005 MACRO domain containing 2 isoform 1 MACROD2 0.005 MACRO domain containing 2 isoform 2 BOD1L 0.005 biorientation of chromosomes in cell division 1-like... ANKRD11 0.005 ankyrin repeat domain 11Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
