
| Name: FAM83A | Sequence: fasta or formatted (434aa) | NCBI GI: 40254997 | |
|
Description: hypothetical protein LOC84985 isoform a
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {367aa} hypothetical protein LOC84985 isoform b | |||
|
Composition:
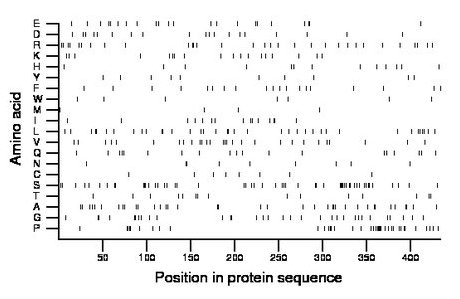
Amino acid Percentage Count Longest homopolymer A alanine 8.5 37 2 C cysteine 1.8 8 1 D aspartate 4.4 19 1 E glutamate 5.1 22 2 F phenylalanine 4.4 19 1 G glycine 7.4 32 2 H histidine 3.0 13 1 I isoleucine 3.5 15 2 K lysine 4.1 18 1 L leucine 8.5 37 2 M methionine 0.9 4 1 N asparagine 2.1 9 2 P proline 8.8 38 5 Q glutamine 4.4 19 1 R arginine 7.1 31 2 S serine 12.2 53 4 T threonine 3.5 15 1 V valine 6.5 28 1 W tryptophan 1.6 7 1 Y tyrosine 2.3 10 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC84985 isoform a FAM83A 0.772 hypothetical protein LOC84985 isoform b FAM83C 0.263 hypothetical protein LOC128876 FAM83H 0.169 FAM83H FAM83F 0.158 hypothetical protein LOC113828 FAM83D 0.155 hypothetical protein LOC81610 FAM83G 0.153 hypothetical protein LOC644815 FAM83B 0.153 hypothetical protein LOC222584 FAM83E 0.140 hypothetical protein LOC54854 LDB3 0.027 LIM domain binding 3 isoform 1 COL3A1 0.022 collagen type III alpha 1 preproprotein LOC100293268 0.021 PREDICTED: hypothetical protein SF3B4 0.021 splicing factor 3b, subunit 4 RERE 0.020 atrophin-1 like protein isoform b RERE 0.020 atrophin-1 like protein isoform a RERE 0.020 atrophin-1 like protein isoform a LDB3 0.020 LIM domain binding 3 isoform 2 ABI3 0.020 NESH protein isoform 2 ABI3 0.020 NESH protein isoform 1 FMN1 0.020 formin 1 ESPN 0.020 espin ZMIZ1 0.020 retinoic acid induced 17 SFPQ 0.020 splicing factor proline/glutamine rich (polypyrimidin... NACA 0.020 nascent polypeptide-associated complex alpha subuni... WDR33 0.020 WD repeat domain 33 isoform 1 TAF4 0.019 TBP-associated factor 4 PRB1 0.019 proline-rich protein BstNI subfamily 1 isoform 1 pre... PRB1 0.019 proline-rich protein BstNI subfamily 1 isoform 2 pre... EN1 0.019 engrailed homeobox 1 LOC100291555 0.019 PREDICTED: hypothetical protein XP_002345954Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
