
| Name: LOC100287767 | Sequence: fasta or formatted (156aa) | NCBI GI: 239742871 | |
|
Description: PREDICTED: hypothetical protein XP_002342668
| Not currently referenced in the text | ||
|
Composition:
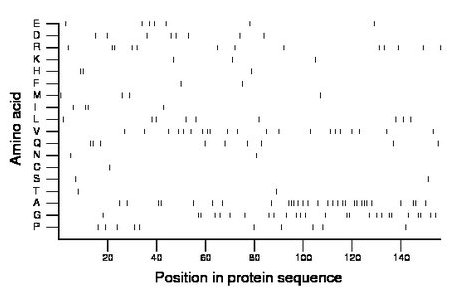
Amino acid Percentage Count Longest homopolymer A alanine 19.2 30 3 C cysteine 0.6 1 1 D aspartate 5.1 8 1 E glutamate 4.5 7 1 F phenylalanine 1.3 2 1 G glycine 16.7 26 2 H histidine 1.9 3 2 I isoleucine 2.6 4 2 K lysine 1.9 3 1 L leucine 5.8 9 1 M methionine 2.6 4 1 N asparagine 1.3 2 1 P proline 6.4 10 1 Q glutamine 5.8 9 2 R arginine 8.3 13 2 S serine 1.3 2 1 T threonine 1.3 2 1 V valine 13.5 21 2 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342668 LOC100291588 1.000 PREDICTED: hypothetical protein XP_002345935 LOC100290285 1.000 PREDICTED: hypothetical protein XP_002346836 HECA 0.060 headcase ARID1B 0.057 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.057 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.057 AT rich interactive domain 1B (SWI1-like) isoform 3 ... KAT2B 0.050 K(lysine) acetyltransferase 2B RNF112 0.050 ring finger protein 112 ELN 0.046 elastin isoform e precursor ELN 0.046 elastin isoform d precursor ELN 0.046 elastin isoform c precursor ELN 0.046 elastin isoform b precursor ELN 0.046 elastin isoform a precursor PHOX2B 0.043 paired-like homeobox 2b RASA1 0.043 RAS p21 protein activator 1 isoform 1 ZIC2 0.043 zinc finger protein of the cerebellum 2 EN1 0.039 engrailed homeobox 1 ZSWIM6 0.039 zinc finger, SWIM-type containing 6 MAZ 0.039 MYC-associated zinc finger protein isoform 2 MAZ 0.039 MYC-associated zinc finger protein isoform 1 LOC728767 0.039 PREDICTED: hypothetical protein LOC728767 0.039 PREDICTED: hypothetical protein FLJ10357 0.039 hypothetical protein LOC55701 TSC22D1 0.036 TSC22 domain family, member 1 isoform 1 ACCN1 0.036 amiloride-sensitive cation channel 1, neuronal isofo... LOC100129410 0.036 PREDICTED: hypothetical protein LOC100129410 0.036 PREDICTED: hypothetical protein LOC100129410 0.036 PREDICTED: hypothetical protein UBE2Z 0.036 ubiquitin-conjugating enzyme E2ZHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
