
| Name: CCK | Sequence: fasta or formatted (115aa) | NCBI GI: 4502605 | |
|
Description: cholecystokinin preproprotein
|
Referenced in: Stomach, Small Intestine, and Colon
| ||
|
Composition:
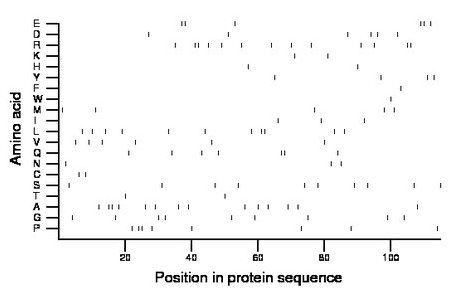
Amino acid Percentage Count Longest homopolymer A alanine 12.2 14 2 C cysteine 1.7 2 1 D aspartate 5.2 6 1 E glutamate 5.2 6 2 F phenylalanine 0.9 1 1 G glycine 7.8 9 1 H histidine 1.7 2 1 I isoleucine 2.6 3 1 K lysine 1.7 2 1 L leucine 9.6 11 2 M methionine 4.3 5 1 N asparagine 2.6 3 1 P proline 7.0 8 2 Q glutamine 6.1 7 2 R arginine 11.3 13 2 S serine 8.7 10 1 T threonine 1.7 2 1 V valine 5.2 6 1 W tryptophan 0.9 1 1 Y tyrosine 3.5 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 cholecystokinin preproprotein GAST 0.048 gastrin preproprotein GLI2 0.034 GLI-Kruppel family member GLI2 FLJ36644 0.034 PREDICTED: hypothetical protein NHSL1 0.029 NHS-like 1 RUSC2 0.019 RUN and SH3 domain containing 2 RUSC2 0.019 RUN and SH3 domain containing 2 SYNM 0.019 desmuslin isoform B SYNM 0.019 desmuslin isoform A GUCY1A2 0.019 guanylate cyclase 1, soluble, alpha 2 C14orf115 0.019 hypothetical protein LOC55237 LOC100132214 0.014 PREDICTED: similar to calcium-promoted Ras inactiva... LOC100132214 0.014 PREDICTED: similar to calcium-promoted Ras inactiva... LOC100132214 0.014 PREDICTED: similar to calcium-promoted Ras inactiva... LOC100132214 0.014 PREDICTED: similar to calcium-promoted Ras inactiva... RASA4 0.014 RAS p21 protein activator 4 isoform 2 RASA4 0.014 RAS p21 protein activator 4 isoform 1 PDCD7 0.014 programmed cell death 7 ANGPTL4 0.014 angiopoietin-like 4 protein isoform b precursor [Hom... YTHDF2 0.014 high glucose-regulated protein 8 C12orf50 0.014 hypothetical protein LOC160419 KDM5C 0.010 jumonji, AT rich interactive domain 1C isoform 2 [H... KDM5C 0.010 jumonji, AT rich interactive domain 1C isoform 1 [H... LOC100294417 0.010 PREDICTED: hypothetical protein NCOR2 0.010 nuclear receptor co-repressor 2 isoform 2 NCOR2 0.010 nuclear receptor co-repressor 2 isoform 1 POLL 0.010 DNA-directed DNA polymerase lambda MED25 0.010 mediator complex subunit 25 LYL1 0.010 lymphoblastic leukemia derived sequence 1 SFRS2 0.010 splicing factor, arginine/serine-rich 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
