
| Name: LOC100133899 | Sequence: fasta or formatted (149aa) | NCBI GI: 169217799 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
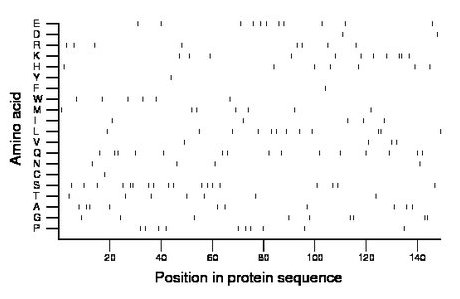
Amino acid Percentage Count Longest homopolymer A alanine 6.7 10 2 C cysteine 0.7 1 1 D aspartate 1.3 2 1 E glutamate 7.4 11 1 F phenylalanine 0.7 1 1 G glycine 6.0 9 2 H histidine 4.7 7 1 I isoleucine 3.4 5 1 K lysine 7.4 11 2 L leucine 8.1 12 2 M methionine 4.7 7 1 N asparagine 2.7 4 1 P proline 6.7 10 1 Q glutamine 10.1 15 2 R arginine 5.4 8 1 S serine 12.1 18 2 T threonine 4.7 7 1 V valine 2.7 4 1 W tryptophan 4.0 6 1 Y tyrosine 0.7 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290382 1.000 PREDICTED: hypothetical protein XP_002347011 LOC100289617 1.000 PREDICTED: hypothetical protein XP_002342956 DLEC1 0.042 deleted in lung and esophageal cancer 1 isoform DLEC... DLEC1 0.042 deleted in lung and esophageal cancer 1 isoform DLEC... MUC16 0.028 mucin 16 RAF1 0.025 v-raf-1 murine leukemia viral oncogene homolog 1 [Hom... CHEK2 0.021 protein kinase CHK2 isoform c CHEK2 0.021 protein kinase CHK2 isoform b CHEK2 0.021 protein kinase CHK2 isoform a MEIS1 0.018 Meis homeobox 1 CYB561D1 0.018 cytochrome b-561 domain containing 1 isoform 5 [Hom... CYB561D1 0.018 cytochrome b-561 domain containing 1 isoform 4 [Hom... CYB561D1 0.018 cytochrome b-561 domain containing 1 isoform 1 [Hom... KIAA0753 0.014 hypothetical protein LOC9851 TAOK3 0.011 TAO kinase 3 RILPL1 0.011 Rab interacting lysosomal protein-like 1 TOX3 0.011 TOX high mobility group box family member 3 isoform... TOX3 0.011 TOX high mobility group box family member 3 isoform... KRT8 0.011 keratin 8 GIGYF2 0.011 GRB10 interacting GYF protein 2 isoform c GIGYF2 0.011 GRB10 interacting GYF protein 2 isoform a GIGYF2 0.011 GRB10 interacting GYF protein 2 isoform b GIGYF2 0.011 GRB10 interacting GYF protein 2 isoform b MLLT4 0.007 myeloid/lymphoid or mixed-lineage leukemia (trithora... BRD4 0.007 bromodomain-containing protein 4 isoform long CENPF 0.007 centromere protein F TRERF1 0.007 transcriptional regulating factor 1 CARD14 0.007 caspase recruitment domain protein 14 isoform 1 [Hom... LAMA1 0.007 laminin, alpha 1 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
