
| Name: RCSD1 | Sequence: fasta or formatted (416aa) | NCBI GI: 217035154 | |
|
Description: RCSD domain containing 1
| Not currently referenced in the text | ||
|
Composition:
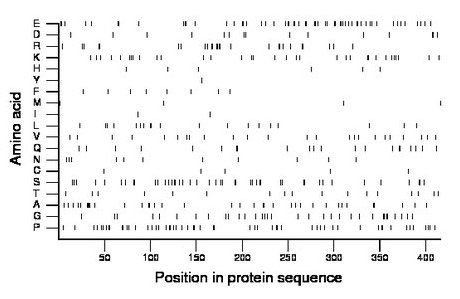
Amino acid Percentage Count Longest homopolymer A alanine 8.7 36 4 C cysteine 1.2 5 1 D aspartate 3.6 15 2 E glutamate 13.5 56 2 F phenylalanine 1.9 8 1 G glycine 7.9 33 2 H histidine 1.4 6 1 I isoleucine 0.5 2 1 K lysine 7.9 33 2 L leucine 4.6 19 2 M methionine 1.0 4 1 N asparagine 2.6 11 1 P proline 13.2 55 2 Q glutamine 5.0 21 2 R arginine 6.2 26 2 S serine 10.6 44 2 T threonine 4.1 17 1 V valine 5.8 24 1 W tryptophan 0.0 0 0 Y tyrosine 0.2 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 RCSD domain containing 1 NEFH 0.066 neurofilament, heavy polypeptide 200kDa RP1L1 0.049 retinitis pigmentosa 1-like 1 RPGR 0.043 retinitis pigmentosa GTPase regulator isoform C [Hom... BAT2 0.040 HLA-B associated transcript-2 FAM21A 0.039 hypothetical protein LOC387680 FAM21B 0.039 hypothetical protein LOC55747 BAT2D1 0.039 HBxAg transactivated protein 2 FSCB 0.038 fibrous sheath CABYR binding protein SMTNL1 0.038 smoothelin-like 1 MYST3 0.036 MYST histone acetyltransferase (monocytic leukemia)... MYST3 0.036 MYST histone acetyltransferase (monocytic leukemia)... MYST3 0.036 MYST histone acetyltransferase (monocytic leukemia)... MAP7D1 0.034 MAP7 domain containing 1 AFF1 0.033 myeloid/lymphoid or mixed-lineage leukemia trithorax ... MLL2 0.033 myeloid/lymphoid or mixed-lineage leukemia 2 PCLO 0.033 piccolo isoform 2 PCLO 0.033 piccolo isoform 1 DNMT1 0.032 DNA (cytosine-5-)-methyltransferase 1 isoform a [Ho... ERICH1 0.032 glutamate-rich 1 TCOF1 0.032 Treacher Collins-Franceschetti syndrome 1 isoform f... TCOF1 0.032 Treacher Collins-Franceschetti syndrome 1 isoform e... TCOF1 0.032 Treacher Collins-Franceschetti syndrome 1 isoform d... TCOF1 0.032 Treacher Collins-Franceschetti syndrome 1 isoform b ... TCOF1 0.032 Treacher Collins-Franceschetti syndrome 1 isoform a ... TCOF1 0.032 Treacher Collins-Franceschetti syndrome 1 isoform c ... PCDH15 0.031 protocadherin 15 isoform CD2-1 precursor LOC100132015 0.031 PREDICTED: similar to testis expressed sequence 13A... BASP1 0.029 brain abundant, membrane attached signal protein 1 [... LOC100132015 0.029 PREDICTED: similar to testis expressed sequence 13A...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
