
| Name: LEO1 | Sequence: fasta or formatted (666aa) | NCBI GI: 20270337 | |
|
Description: Leo1, Paf1/RNA polymerase II complex component, homolog
|
Referenced in: RNA Polymerase and General Transcription Factors
| ||
|
Composition:
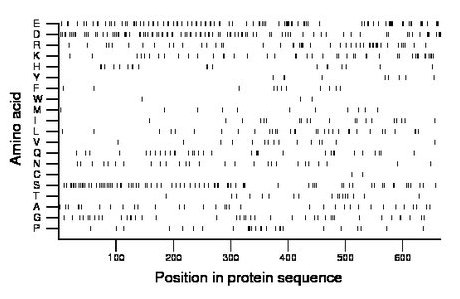
Amino acid Percentage Count Longest homopolymer A alanine 5.3 35 2 C cysteine 0.3 2 1 D aspartate 14.4 96 3 E glutamate 14.7 98 4 F phenylalanine 1.7 11 1 G glycine 6.2 41 2 H histidine 2.9 19 2 I isoleucine 3.0 20 1 K lysine 6.6 44 2 L leucine 4.8 32 2 M methionine 1.8 12 1 N asparagine 4.1 27 1 P proline 3.9 26 2 Q glutamine 5.1 34 2 R arginine 7.1 47 2 S serine 11.7 78 2 T threonine 2.6 17 1 V valine 2.3 15 1 W tryptophan 0.5 3 1 Y tyrosine 1.4 9 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 Leo1, Paf1/RNA polymerase II complex component, homo... DSPP 0.084 dentin sialophosphoprotein preproprotein IWS1 0.071 IWS1 homolog ACRC 0.055 ACRC protein RPGR 0.049 retinitis pigmentosa GTPase regulator isoform C [Hom... RP1L1 0.049 retinitis pigmentosa 1-like 1 DMP1 0.045 dentin matrix acidic phosphoprotein 1 isoform 1 precu... DMP1 0.045 dentin matrix acidic phosphoprotein 1 isoform 2 pre... MDN1 0.044 MDN1, midasin homolog HTATSF1 0.042 HIV-1 Tat specific factor 1 HTATSF1 0.042 HIV-1 Tat specific factor 1 RBM25 0.040 RNA binding motif protein 25 CALD1 0.039 caldesmon 1 isoform 1 ATRX 0.037 transcriptional regulator ATRX isoform 2 ATRX 0.037 transcriptional regulator ATRX isoform 1 HRC 0.036 histidine rich calcium binding protein precursor [Hom... LOC100286959 0.036 PREDICTED: hypothetical protein XP_002343921 RTF1 0.035 Paf1/RNA polymerase II complex component FLG 0.034 filaggrin SETD1B 0.034 SET domain containing 1B CYLC1 0.033 cylicin, basic protein of sperm head cytoskeleton 1... TCHH 0.033 trichohyalin ZC3H13 0.032 zinc finger CCCH-type containing 13 GOLIM4 0.032 golgi integral membrane protein 4 YTHDC1 0.032 splicing factor YT521-B isoform 2 YTHDC1 0.032 splicing factor YT521-B isoform 1 SPARCL1 0.030 SPARC-like 1 SPARCL1 0.030 SPARC-like 1 SFRS12 0.030 splicing factor, arginine/serine-rich 12 isoform a ... SFRS12 0.030 splicing factor, arginine/serine-rich 12 isoform b [...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
