
| Name: POU3F2 | Sequence: fasta or formatted (443aa) | NCBI GI: 51702521 | |
|
Description: POU domain, class 3, transcription factor 2
|
Referenced in: Protein Composition and Structure
| ||
|
Composition:
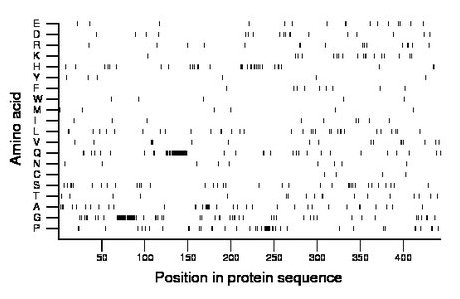
Amino acid Percentage Count Longest homopolymer A alanine 8.1 36 5 C cysteine 0.9 4 1 D aspartate 4.1 18 2 E glutamate 3.6 16 2 F phenylalanine 2.0 9 1 G glycine 14.2 63 21 H histidine 7.4 33 3 I isoleucine 2.0 9 1 K lysine 4.1 18 2 L leucine 7.4 33 2 M methionine 1.4 6 1 N asparagine 2.0 9 1 P proline 9.5 42 7 Q glutamine 11.3 50 21 R arginine 3.6 16 2 S serine 7.9 35 3 T threonine 4.1 18 2 V valine 3.8 17 3 W tryptophan 1.1 5 1 Y tyrosine 1.4 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 POU domain, class 3, transcription factor 2 POU3F3 0.467 POU class 3 homeobox 3 POU3F4 0.401 POU domain, class 3, transcription factor 4 POU3F1 0.367 POU domain, class 3, transcription factor 1 POU2F1 0.208 POU class 2 homeobox 1 POU2F2 0.195 POU domain, class 2, transcription factor 2 POU2F3 0.194 POU transcription factor POU4F2 0.179 Brn3b POU domain transcription factor POU5F1 0.177 POU domain, class 5, transcription factor 1 isoform ... POU5F1 0.177 POU domain, class 5, transcription factor 1 isoform... POU5F1B 0.172 POU class 5 homeobox 1B POU1F1 0.172 pituitary specific transcription factor 1 isoform alp... POU1F1 0.171 pituitary specific transcription factor 1 isoform b... POU4F3 0.147 POU class 4 homeobox 3 POU4F1 0.146 POU domain, class 4, transcription factor 1 POU5F2 0.127 POU domain class 5, transcription factor 2 POU6F1 0.120 POU class 6 homeobox 1 POU6F2 0.087 POU domain, class 6, transcription factor 2 FOXJ2 0.046 forkhead box J2 ATXN2 0.044 ataxin 2 ATN1 0.044 atrophin-1 ATN1 0.044 atrophin-1 MAFA 0.042 v-maf musculoaponeurotic fibrosarcoma oncogene homol... MN1 0.039 meningioma 1 AR 0.038 androgen receptor isoform 1 SFPQ 0.037 splicing factor proline/glutamine rich (polypyrimidin... SMARCA2 0.037 SWI/SNF-related matrix-associated actin-dependent re... SMARCA2 0.037 SWI/SNF-related matrix-associated actin-dependent re... NCOA6 0.037 nuclear receptor coactivator 6 FAM155A 0.037 family with sequence similarity 155, member A [Homo...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
