
| Name: LOC100133981 | Sequence: fasta or formatted (394aa) | NCBI GI: 239755446 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
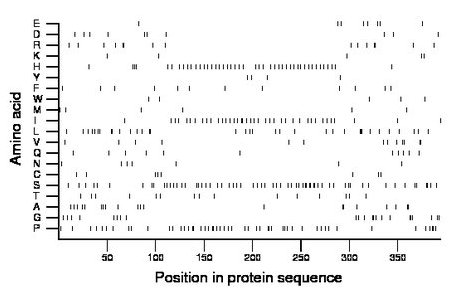
Amino acid Percentage Count Longest homopolymer A alanine 5.1 20 2 C cysteine 2.0 8 1 D aspartate 2.5 10 1 E glutamate 2.0 8 1 F phenylalanine 3.0 12 1 G glycine 5.6 22 3 H histidine 11.4 45 2 I isoleucine 9.9 39 2 K lysine 1.3 5 1 L leucine 8.9 35 3 M methionine 1.5 6 1 N asparagine 1.8 7 1 P proline 12.9 51 2 Q glutamine 2.8 11 1 R arginine 3.8 15 2 S serine 16.0 63 2 T threonine 3.8 15 1 V valine 3.3 13 2 W tryptophan 1.3 5 1 Y tyrosine 1.0 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100133439 0.732 PREDICTED: hypothetical protein LOC100131525 0.260 PREDICTED: hypothetical protein LOC100291393 0.253 PREDICTED: hypothetical protein XP_002347924 LOC100288473 0.253 PREDICTED: hypothetical protein XP_002343336 LOC100288786 0.235 PREDICTED: hypothetical protein XP_002343819 LOC100132614 0.233 PREDICTED: hypothetical protein, partial LOC100293557 0.222 PREDICTED: hypothetical protein LOC100133553 0.054 PREDICTED: hypothetical protein, partial LOC100133173 0.054 PREDICTED: hypothetical protein LOC100134060 0.046 PREDICTED: hypothetical protein LOC100134332 0.041 PREDICTED: hypothetical protein, partial LOC100288317 0.041 PREDICTED: hypothetical protein XP_002342505 LOC100133515 0.040 PREDICTED: hypothetical protein PRR21 0.040 proline rich 21 SF3A2 0.038 splicing factor 3a, subunit 2 LOC100132983 0.038 PREDICTED: hypothetical protein LOC100290755 0.036 PREDICTED: hypothetical protein XP_002346496 LOC100131203 0.036 PREDICTED: hypothetical protein LOC100129802 0.036 PREDICTED: hypothetical protein, partial LOC100128719 0.035 PREDICTED: hypothetical protein, partial LOC100128719 0.035 PREDICTED: hypothetical protein, partial LOC100134325 0.035 PREDICTED: hypothetical protein LOC100129802 0.033 PREDICTED: hypothetical protein LOC284297 0.032 hypothetical protein LOC284297 LOC100128267 0.028 PREDICTED: hypothetical protein, partial LOC100133741 0.027 PREDICTED: hypothetical protein LOC100129642 0.027 PREDICTED: hypothetical protein LOC100290974 0.027 PREDICTED: hypothetical protein XP_002346910 PHLDA1 0.027 pleckstrin homology-like domain, family A, member 1 ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
